Separating hyperplane tutorial¶
This is a very introductory tutorial, showing how a classification task (in this case, deciding whether people are sumo wrestlers or basketball players, based on their height and weight) can be viewed as drawing a decision boundary in a feature space. It shows how to plot the data, calculate the weights of a simple linear classifier, and see how the resulting classifier carves up the feature space into two categories.
Note
This tutorial was originally written by Rajeev Raizada for Matlab and was ported to Python by the PyMVPA authors. The original Matlab code is available from: http://www.dartmouth.edu/~raj/Matlab/fMRI/classification_plane_tutorial.m
Let’s look at a toy example: classifying people as either sumo wrestlers or basketball players, depending on their height and weight. Let’s call the x-axis height and the y-axis weight
sumo_wrestlers_height = [ 4, 2, 2, 3, 4 ]
sumo_wrestlers_weight = [ 8, 6, 2, 5, 7 ]
basketball_players_height = [ 3, 4, 5, 5, 3 ]
basketball_players_weight = [ 2, 5, 3, 7, 3 ]
Let’s plot this.
import pylab as pl
pl.plot(sumo_wrestlers_height, sumo_wrestlers_weight, 'ro',
linewidth=2, label="Sumo wrestlers")
pl.plot(basketball_players_height, basketball_players_weight, 'bx',
linewidth=2, label="Basketball players")
pl.xlim(0, 6)
pl.ylim(0, 10)
pl.xlabel('Height')
pl.ylabel('Weight')
pl.legend()
Let’s stack up the sumo data on top of the basketball players data.
import numpy as np
# transpose to have observations along the first axis
sumo_data = np.vstack((sumo_wrestlers_height,
sumo_wrestlers_weight)).T
# same for the baskball data
basketball_data = np.vstack((basketball_players_height,
basketball_players_weight)).T
# now stack them all together
all_data = np.vstack((sumo_data, basketball_data))
In order to be able to train a classifier on the input vectors, we need to know
what the desired output categories are for each one. Let’s set this to be +1
for sumo wrestlers, and -1 for basketball players.
# creates: [ 1, 1, 1, 1, 1, -1, -1, -1, -1, -1]
all_desired_output = np.repeat([1, -1], 5)
We want to find a linear decision boundary,
i.e. simply a straight line, such that all the data points
on one side of the line get classified as sumo wrestlers,
i.e. get mapped onto the desired output of +1,
and all the data points on the other side get classified
as basketball players, i.e. get mapped onto the desired output of -1.
The equation for a straight line has this form:
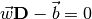
were 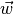 is a weight vector,
is a weight vector, 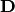 is the data matrix,
and
is the data matrix,
and 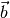 is the offset of the dataset form the origin, or the bias.
We’re not so interested for now in
is the offset of the dataset form the origin, or the bias.
We’re not so interested for now in 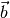 ,
so we can get rid of that by subtracting the mean from our data to get
,
so we can get rid of that by subtracting the mean from our data to get
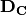 the per-column (i.e. variable) demeaned data that is now
centered around the origin.
the per-column (i.e. variable) demeaned data that is now
centered around the origin.
zero_meaned_data = all_data - all_data.mean(axis=0)
Now, having gotten rid of that annoying bias term, we want to find a weight vector which gives us the best solution that we can find to this equation:
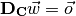
were 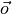 is the desired output, or the class labels. But, there is
no such perfect set of weights. We can only get a best fit, such that
is the desired output, or the class labels. But, there is
no such perfect set of weights. We can only get a best fit, such that
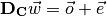
where the error term 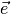 is as small as possible.
is as small as possible.
Note that our equation
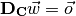
has exactly the same form as the equation from the tutorial code in http://www.dartmouth.edu/~raj/Matlab/fMRI/design_matrix_tutorial.m which is:
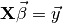
where 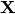 was the design matrix,
was the design matrix, 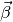 the
sensitivity vector, and
the
sensitivity vector, and 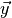 the voxel response.
the voxel response.
The way we solve the equation is exactly the same, too. If we could find a matrix-inverse of the data matrix, then we could pre-multiply both sides by that inverse, and that would give us the weights:
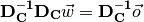
The 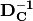 and
and 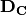 terms on the left
would cancel each other out, and we would be left with:
terms on the left
would cancel each other out, and we would be left with:
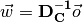
However, unfortunately there will in general not exist any matrix-inverse of
the data matrix 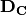 . Only square matrices have inverses,
and not even all of them do. Luckily, however, we can use something that plays
a similar role, called a pseudo-inverse. In Numpy, this is given by the command
. Only square matrices have inverses,
and not even all of them do. Luckily, however, we can use something that plays
a similar role, called a pseudo-inverse. In Numpy, this is given by the command
pinv. The pseudo-inverse won’t give us a perfect solution to the above
equation but it will give us the best approximate solution, which is what we
want.
So, instead of
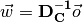
we have this equation:
# compute pseudo-inverse as a matrix
pinv = np.linalg.pinv(np.mat(zero_meaned_data))
# column-vector of observations
y = all_desired_output[np.newaxis].T
weights = pinv * y
Let’s have a look at how these weights carve up the input space A useful command for making grids of points which span a particular 2D space is called “meshgrid”
gridspec = np.linspace(-4, 4, 20)
input_space_X, input_space_Y = np.meshgrid(gridspec, gridspec)
# for the rest it is easier to have `weights` as a simple array, instead
# of a matrix
weights = weights.A
weighted_output_Z = input_space_X * weights[0] + input_space_Y * weights[1]
The weighted output gets turned into the category-decision +1
if it is greater than 0, and -1 if it is less than zero.
Let’s plot the decision surface color-coded and then plot the zero-meaned
sumo and basketball data on top.
pl.figure()
pl.pcolor(input_space_X, input_space_Y, weighted_output_Z,
cmap=pl.cm.Spectral)
pl.plot(zero_meaned_data[all_desired_output == 1, 0],
zero_meaned_data[all_desired_output == 1, 1],
'ro', linewidth=2, label="Sumo wrestlers")
pl.plot(zero_meaned_data[all_desired_output == -1, 0],
zero_meaned_data[all_desired_output == -1, 1],
'bx', linewidth=2, label="Basketball players")
pl.xlim(-4, 4)
pl.ylim(-4, 4)
pl.colorbar()
pl.xlabel('Demeaned height')
pl.ylabel('Demeaned weight')
pl.title('Decision output')
pl.legend()
from mvpa2.base import cfg
See also
The full source code of this example is included in the PyMVPA source distribution (doc/examples/hyperplane_demo.py).




